Chapter 4 Co-Variates and Raster Files
4.1 CHELSA global downscaled climate data set
Let me know if you want to see the technical process of modifying raster files using the NJ shapefile.
# Example: Reading Raster Files from Source folder to R
```{r setup, include=FALSE}
library(raster)
# Read in Covariates from Folder name (e.g. chelsa)
fileDir <- "chelsa"
files <- list.files(fileDir, pattern = '.tif$', full.names = TRUE)
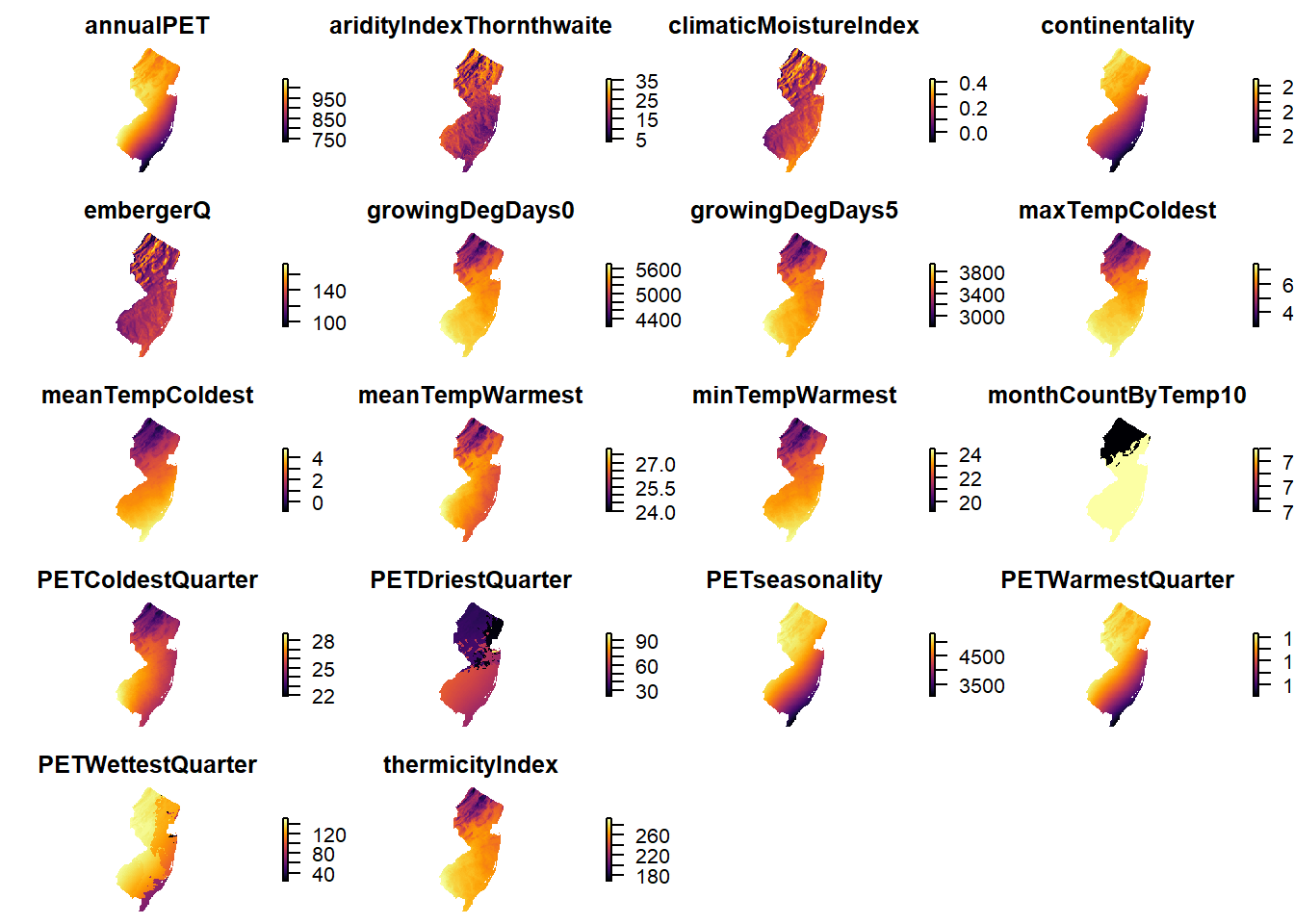
```## [1] "annualPET" "aridityIndexThornthwaite"
## [3] "climaticMoistureIndex" "continentality"
## [5] "embergerQ" "growingDegDays0"
## [7] "growingDegDays5" "maxTempColdest"
## [9] "meanTempColdest" "meanTempWarmest"
## [11] "minTempWarmest" "monthCountByTemp10"
## [13] "PETColdestQuarter" "PETDriestQuarter"
## [15] "PETseasonality" "PETWarmestQuarter"
## [17] "PETWettestQuarter" "thermicityIndex"